
Research
Interests:
Statistical mechanics of complex fluids.
For almost two decades our efforts have been broadly focused on "complex fluids", from both analytical and computational points of view. Examples of such systems include: micellized surfactant solutions and microemulsions; lipid bilayers; colloidal suspensions; polymer solutions, including polyelectrolytes; interfacial and surface thin films; and liquid crystals. In all cases the systems involve a mesoscopic length scale -- nanometers, typically -- intermediate between the microscopic and macroscopic. Correspondingly, it is almost always inappropriate to describe them via use of a molecular, atomistic, hamiltonian. Instead, the various energies and entropy terms are best treated phenomenologically, in order to address the basic questions involving structural evolution and phase transitions on mesoscopic scales. Our work up through 1996, along with a critical perspective of the field, is reviewed in the Centennial issue (volume 100) of the Journal of Physical Chemistry: W. M. Gelbart and A. Ben-Shaul, "The 'New' Science of Complex Fluids", pp. 13169-89. More recent investigations, featuring spontaneous pattern formation in nanoparticle dispersions, are described in W. M. Gelbart, R. P. Sear, J. R. Heath and S. Chaney, "Array Formation in Nano-colloids: Theory and Experiment in 2D", Faraday Discussions 112, 299-307 (1999).
Life cycles of viruses: DNA condensation, complexation, and encapsidation
|
As
a natural outgrowth of our longstanding interest in the statistical
mechanics of complex fluids, we have begun to concentrate recently
on a wide range of problems involving biological consequences of
the mesoscopic properties of DNA. Even as it is the basic repository
of genetic information, DNA is also "just" a linear polyelectrolyte
and 1D elastic object whose consequent physical properties are of
fundamental importance to all living systems. DNA in most bacterial
viruses, for example, is strongly condensed, i.e., its density is
comparable to that of crystalline DNA; furthermore, it is confined
in protein capsids whose dimensions are orders of magnitude smaller
than its length (see Fig. 1). We are interested in how DNA is packaged
in these viral capsids (Fig. 2) and how its state of stress determines
the way it is injected into a bacterial cell to begin the infection
process. In the case of most animal viruses, like polio, flu and
HIV, on the other hand, infection is initiated by passage of the
entire viral particle -- protein capsid and all -- through the plasma
membrane into the cell cytoplasm. We study this process -- endocytosis
-- from a physical point of view, i.e., a generic colloidal particle
passing through a molecular bilayer; phenomenological connections
to specific biological systems are developed by treating, for example,
the effect of receptor proteins on membrane elasticity and particle/membrane
adhesion energy, etc. We also investigate the structure and dynamics
of nucleosomes, the basic building blocks of chromosomes in which
DNA is complexed with globular aggregates of cationic proteins (the
histones). In another set of problems we are attempting to understand
how -- and why -- icosahedral symmetry plays such a fundamental
role in the self-assembly of viral protein capsids (Fig. 3).
|
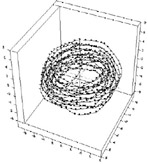
Fig.
1 shows a typical configuration of a 694 bead-chain which has
been loaded into a capsid of diameter 16 (in units of bead size);
the chain persistence length is 32, and the pair attractive energy
between beads is 0.5 kBT.
|
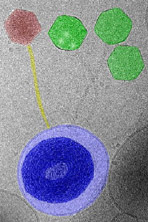
Fig.
2. In this cryo-TEM image, three T5 bacterial viruses (green)
have injected their genomes into a lipid vesicle that has been
reconstituted with the receptor proteins for T5 -- they have mistaken
this vesicle for a bacterial cell host; a fourth viral particle
(red) is about to do the same.
|
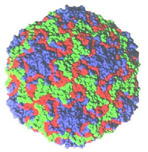
Fig.
3. Image-reconstructed icosahedral structure of the human rhinovirus
protein capsid.
|
Selected Publications:
- Kindt, James; Tzlil, Shelly; Ben-Shaul, Avinoam; Gelbart, William M., "DNA Packaging and Ejection Forces in Bacteriophage", Proc. Natl. Acad. Sci. U. S. A. 98(24), 13671-13674 (2001).
- Schiessel, Helmut; Gelbart, William M.; Bruinsma, Robijn, "DNA Folding: Structural and Mechanical Properties of the Two-Angle Model for Chromatin",Biophys. J. 80(4), 1940-1956 (2001).
- Borukhov, Itamar; Bruinsma, Robijn F.; Gelbart, William M.; Liu, Andrea J.," Elastically Driven Linker Aggregation between Two Semiflexible Polyelectrolytes", Phys. Rev. Lett. 86(10), 2182-2185 (2001).
- Schiessel, Helmut; Widom, Jonathan; Bruinsma, Robijn F.; Gelbart, William M., "Polymer Repatation and Nucleosome Repositioning", Phys. Rev. Lett. 86(19), 4414-4417 (2001).
- Gelbart, William M.; Bruinsma, Robijn F.; Pincus, Philip A.; Parsegian, V. Adrian, "DNA-Inspired Electrostatics", Physics Today, 38-44 (September 2000).
Biography:
Dr. Gelbart received his B.S. (1967) at Harvard University and his M.S. (1968) and Ph.D. (1970) at the University of Chicago. After an NSF/NATO postdoctoral fellowship at the University of Paris he joined the University of California as an Assistant Professor at Berkeley in 1972 before moving to UCLA in 1975, where he has been Professor of Chemistry and Biochemistry since 1979. Dr. Gelbart's research interests have evolved from molecular quantum mechanics in the gas phase to optical properties of simple and critical fluids, to liquid crystals and complex fluids, and "finally" to biophysical problems. His theoretical contributions have been recognized by Sloan (1974) and Guggenheim (1998) Fellowships, the Lennard-Jones Medal of the British Royal Society (1991), the ACS "Liquids" (Hildebrand) Prize (2001), and by endowed lectureships throughout the States and Europe. In 1996 he won the UCLA Distinguished Teaching Award.
|


