
|
UCLA
Departments of Chemistry & Biochemistry, Biological Chemistry; UCLA-DOE Laboratory of Structural Biology & Molecular Medicine
201 Boyer Hall
Box 951570
Los Angeles, CA 90095-1570
|
Tel: (310) 825-3754
Fax: (310) 206-3914
Email: david@mbi.ucla.edu
Website: www.doe-mbi.ucla.edu/People/Eisenberg/
| |
|
Research
Interests:
|
The long term goal is to understand and manipulate the metabolism
of cells through the interactions of their constituent proteins.
One goal is to be able to infer functional linkages of proteins,
on the basis of genome sequences and protein expression data.
Computational methods have been developed for establishing
these relationships, including the phylogenetic profile and
Rosetta Stone methods. To benchmark these computational
methods, a large Database of Interacting Proteins has been
built up. X-ray crystallography remains a powerful tool for
exploring protein structure and interactions. Our X-ray projects
are of two types. The first are on amyloids and prions, pathologically
interacting proteins. The goal is to understand the structures
that underlie the pathologies. The other projects are on the
structural biology of Mycobacterium tuberculosis, as part
of the TB Structural Genomics Consortium.
| |
To
the right is the crystal structure of glutamine
sythetase studied by our X-ray crystollagraphic methods.
|
|
|
3D
domain swapping is a mechanism for proteins to form oligomers
by exchanging identical domains. To date, there are more than
30 domain-swapped proteins with reported structures. Here, we
have chosen bovine pancreatic ribonuclease (RNase A) to study
3D domain swapping. |

|
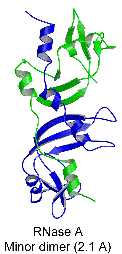
When concentrated in mild acid solutions, bovine pancreatic
ribonuclease (RNase A) forms long-lived oligomers including
two types of dimer, two types of trimer, and higher oligomers.
We have determined the structures of the two dimers and one
trimer and built a model for the other trimer, shown in the
picture. In the dimer or trimer, one component is more than
the other, thus called major dimer/trimer and minor dimer/trimer,
respectively. The major dimeric component forms by a swapping
of the C-terminal ß-strands between the monomers, whereas
the minor dimeric component forms by swapping the N-terminal
alpha helices of the monomers.
|
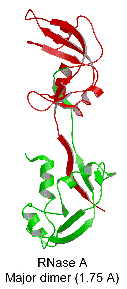
 |
 |
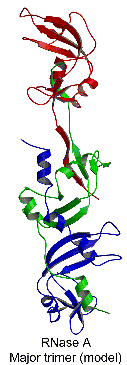
Based on these structures, a linear model for the RNase A
major trimer was proposed to form from a central molecule
that simultaneously swaps its N-terminal helix with a second
RNase A molecule and its C-terminal strand with a third molecule.
Studies by dissociation are consistent with this model: the
major trimer dissociates into both the major and the minor
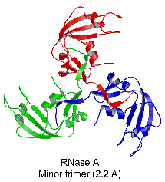
dimers, as well as monomers. The minor trimer is cyclic, formed
from three monomers that each swap their C-terminal ß-strand
into an identical molecule, as shown by the crystal structure.
This study thus expands the variety of domain-swapped oligomers
by revealing the first example of a protein that swaps both
N- and C-termini and that can form both a linear and a cyclic
domain-swapped oligomer.
|
|
As part of a structural genomics pilot project, we have solved
the crystal structures of Sm-like archaeal proteins ("SmAPs")
from the hyperthermophilic archaea Pyrobaculum aerophilum
(PAE) and Methanobacterium thermautotrophicum (MTH), and found
that these novel structures provide insights into the central
architecture of small nuclear ribonucleoproteins (snRNPs).
By forming the cores of snRNP assemblies, eukaryotic Sm and
Lsm proteins are key components of several RNA-processing
machines, including the 5 x 107 Da spliceosome.
|
 |
We determined the PAE SmAP1 crystal
structure to 1.75 A-resolution by MAD phasing, and found that
SmAP1 monomers assemble into a heptameric ring perforated by
a cationic pore (Fig. 1, to the left). In addition to providing
direct evidence for such an assembly in eukaryotic snRNPs, our
results (i) show that SmAP homodimers are structurally similar
to human Sm heterodimers; (ii) show that SmAP is a member of
a phylogenetically well-conserved module that likely evolved
into modern Sm proteins via gene duplication; and (iii) offer
an unconventional model of SmAP1 bound to single stranded RNA
that explains the specificity of Sm binding sites found in small
nuclear RNAs. The pronounced electrostatic asymmetry of the
heptameric disk would impart directionality to putative SmAP-RNA
interactions. |
| The refined PAE SmAP1 model was used
to solve the MTH SmAP1 structure to 1.85 A by molecular replacement.
As expected, PAE and MTH heptamers are structurally quite similar
(Fig. 2, to the right). These results provide an atomic resolution
glimpse into the probable structures of eukaryotic snRNPs, and
raise a number of interesting questions (which we are currently
exploring) regarding the existence of snRNP-like particles in
archaeal species. Preliminary biochemical investigation of SmAP
suggests that it is dissimilar to human Sm proteins in a number
of ways. |
|
Selected
Publications:
- Landgraf R, Xenarios I, Eisenberg D, "Three-dimensional
cluster analysis identifies interfaces and functional residue
clusters in proteins", J Mol Biol. 307(5),
1487-502 (2001).
- Marcotte EM, Xenarios I, Eisenberg D, "Mining literature
for protein-protein interactions", Bioinformatics
17(4), 359-63 (2001).
- Gill HS, Eisenberg D, "The crystal structure of phosphinothricin
in the active site of glutamine synthetase illuminates the mechanism
of enzymatic inhibition", Biochemistry 40(7),
1903-12 (2001).
- Mura C, Cascio D, Sawaya MR, Eisenberg DS, "The crystal
structure of a heptameric archaeal Sm protein: Implications for
the eukaryotic snRNP core", Proc Natl Acad Sci (USA)
98(10), 5532-7 (2001).
- Liu Y, Gotte G, Libonati M, Eisenberg D, "A domain-swapped
RNase A dimer with implications for amyloid formation", Nat
Struct Biol. 8(3), 211-214 (2001).
Dr.
Eisenberg's complete list of publications
Biography:
David
Eisenberg is currently a Professor of Chemistry & Biochemistry,
as well as the Director of the DOE Lab of Structural Biology &
Molecular Medicines at UCLA. Before he assumed his role in the UCLA
community Mr. Eisenberg received an A.B. in Biochemical Science
from Harvard College and preceded to Oxford University where he
completed his D.Phil. in Theoretical Chemistry. After completing
two Postdoctorales, one at Princeton University regarding water
and hydrogen bonding and the other at the California Institute of
Technology on protein crystallography, he joined the faculty at
UCLA. Currently his research involves the use of x-ray crystallography
and the long-term goal of understanding and manipulating the metabolism
of cells through the interactions of their constituent proteins.
Throughout his career Mr. Eisenberg has published over 200 papers,
holds several patents and has presented over 10 lectureships through
the University of Michigan, Purdue, Harvard to name a few. Of his
many awards some highlights include; Alfred P. Sloan Fellow, UCLA
Distinguished Teaching Award, John Simon Guggenheim Fellow, UCLA
Faculty Research Lectureship, National Academy of Sciences Member,
American Academy of Arts and Sciences Member.
|
|


